Project 1. Cellular dynamics during fungal invasion of host cells
To cause disease, the blast fungus undergoes a series of developmental stages (Khang and Valent, 2010).
To cause disease, the blast fungus undergoes a series of developmental stages (Khang and Valent, 2010).
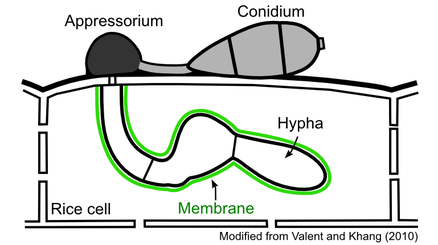
On the rice leaf surface, a fungal conidium germinates to form a germ tube, which is then differentiated into a specialized penetration cell called an appressorium. The matured appressorium uses turgor pressure to penetrate into the epidermal rice cell. Once entering the cell, the fungus produces biotrophic hyphae that grow inside the live host cell. These hyphae remain separated from the host cytoplasm by a membrane that surrounds them.
We study cellular dynamics during fungal invasion of rice cells. We aim to identify key cellular processes that are required for disease development and to determine the genes that control such processes. Our most recent strategies involve live-cell confocal microscopy of fluorescently labeled fungus and rice strains, quantitative image analysis, and molecular genetics tools.
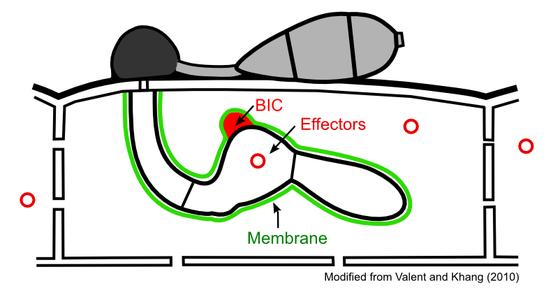
Project 2. Effector Proteins
All classes of pathogens, such as fungi, oomycetes, and bacteria, secrete effector proteins that facilitate colonization by modifying host cell structure and function. Some effectors (avirulence effectors) also trigger host immune responses and block colonization. Thus, effectors are increasingly recognized as a key molecule for the study of microbial pathogenicity and innate host immunity.
We study effectors that the blast fungus produces (Khang et al., 2010; Valent and Khang, 2010). Effectors secreted by hyphae enter the host cytoplasm across the surrounding membrane. The translocated effectors move into uninvaded neighbor cells, presumably preparing host cells before invasion.
We are interested in understanding how blast effectors are expressed, how they are targeted into various subcellular compartments, and what their host targets are. Our current project focuses on mechanisms of effector delivery into the host cytoplasm. We aim to identify host translocation motifs and to characterize biotrophic interfacial complexes (BICs) that are proposed to mediate effector delivery. We use a multidisciplinary approach that includes confocal microscopy, electron microscopy, and molecular genetics tools, as well as developing and applying new technologies.
Project 3. Interfacial Membrane
Diverse fungi form intimate biotrophic associations with plant cells. A central feature of such associations is the presence of a membrane that surrounds intracellular fungal cells. The purpose that this membrane serves is yet unknown. It could be beneficial to either the fungi or plants, detrimental to either, or have a neutral interaction with one or both. There are two possible scenarios for these interactions. One is that the fungus tricks the host cell to produce the membrane in order to protect itself from host defense responses; another is that the plant produces the membrane in an attempt to mechanically halt the growth of the fungus and/or stop effector translocation. When the rice blast fungus invades live rice cells, it differentiates from the initial filamentous primary hypha into bulbous invasive hyphae. These hyphal cells are surrounded by a membrane termed as the extra-invasive hyphal membrane (EIHM). We study the origin, structure, dynamics, and function of the EIHM. Since the EIHM is formed during successful infection of rice cells, our research may offer new insight into possible modes of disease control.